Help for using DeepHomo2.0 server
1. About the DeepHomo2.0
The DeepHomo2.0 is a deep learning-based model for accurate prediction of residue-residue contacts across homo-oligomeric protein interfaces by integrating sequential and structural information of monomers.
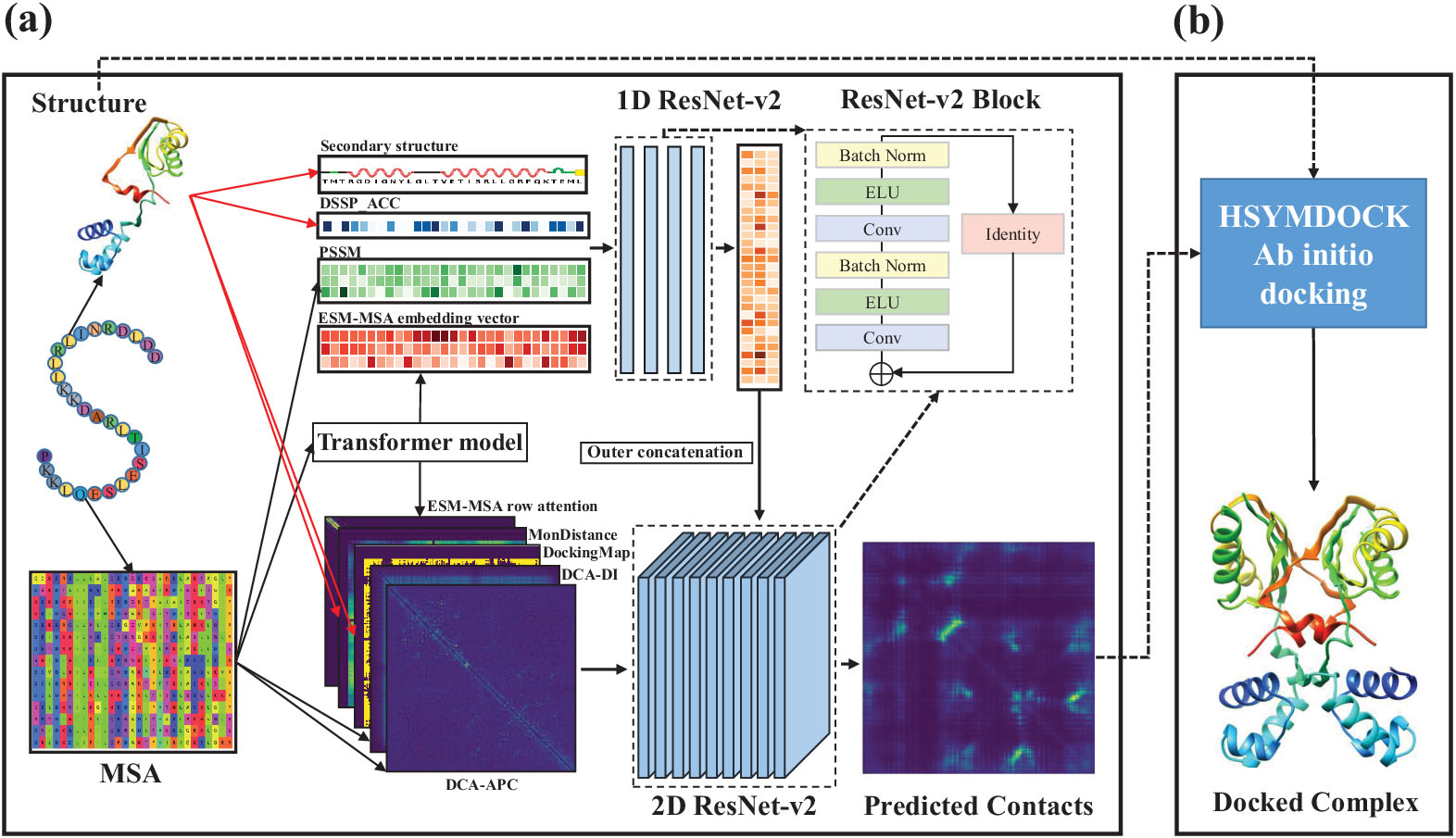
2. How to provide input for the monomer.
Users need to provide the 3D structure of the monomer which can be a experimental structrue from the PDB or a predicted model by a third party approach like I-TASSER. The DeepHomo2.0 server accepts two types of input for the monomer:
- Upload your pdb file in PDB format.
- Copy and paste your pdb file in plain text.
Only ONE type of input is needed for the subunit molecule. If more than one types of input is provided, the first one will be used.
3. How to obtain your DeepHomo2.0 results.
Once users submit your job, they will be redirected to a status web page showing the status of the job. The status page is automatically refreshed every 10 seconds until the job is finished. Users have three ways to obtain their prediction results.- Keep the status page open until it shows the prediction results when the results is ready.
- Bookmark the status page and come back later to check the prediction results.
- Wait for the email notification if users provide a valid email address at the time of job submission.
- The monomer pdb file, monomer sequence, predicted contacts.
- Or a compressed package of the above three files.
As the top 10 contacts are normally deemed as the most important ones, the result page provides an interactive view of the top 10 contacts using the NGL viewer, where two identical momomers are shown side-by-side, displaying the two corresponding residues of a contact. Users can choose to view any of the top 10 contacts or all together by different representations and styles.
The page also gives a summary of the rankings and residue pairs for the top 10 contacts. The predicted score ranges from 0.0 to 1.0. The higher a predicted score is, the more likely to be in contact the residue pairs are.
NOTE: It is recommended that users download their prediction results as soon as possible after the job is done, as the job results will only be stored on our server for two weeks.4. Contact-assisted docking (Optional).
This step is optional. If users want to obtain the homo-dimeric structure of monomers, they need to obtain the standalone HSYMDOCK package at http://huanglab.phys.hust.edu.cn/hsymdock/.Then, users may run a protein-protein docking job using the predicted contacts as restraints like this.
awk '{if(NR>1&&NR<=6)print $2,$4,$6}' monomer_contacts.out > top5.txt
modcheck monomer.pdb > monomer_check.pdb
clean_pdb monomer_check.pdb monomer_clean.pdb
chdock monomer_clean.pdb monomer_clean.pdb -cont top5.txt -out CHdock.out
With the docking output, users can generate 10 complex structures using the following command.
compcn CHdock.out output.pdb -nmax 10
where the "output.pdb" contains 10 complex structures in NMR style,
though users can adjust the number of generated complex structures
by setting the value of "-nmax" option.