Software, Servers and Databases
 HDOCK Server
-- Protein-protein and protein-DNA/RNA docking based on a hybrid algorithm of template-based modeling and ab initio free docking.
HDOCK Server
-- Protein-protein and protein-DNA/RNA docking based on a hybrid algorithm of template-based modeling and ab initio free docking.
 MODPEP Server
-- Fast conformer ensemble generator of protein-bound peptides.
MODPEP Server
-- Fast conformer ensemble generator of protein-bound peptides.
Click HERE to download the training data set.
Click HERE to download the test data set. HPepDock
--
A hierarchical flexible peptide docking approach by fast
conformational modeling and orientational sampling of peptides.
HPepDock
--
A hierarchical flexible peptide docking approach by fast
conformational modeling and orientational sampling of peptides.
Click HERE to access the web server.
 HSYMDOCK
--
A Hierarchical SYMmetric DOCKing protocol for protein homo-oligomers with Cn or Dn symmetry
from a subunit molecule.
HSYMDOCK
--
A Hierarchical SYMmetric DOCKing protocol for protein homo-oligomers with Cn or Dn symmetry
from a subunit molecule.
Click HERE to access the web server.
 FTALIGN
--
Fast topology-independent and global structure alignment through an FFT-based algorithm.
FTALIGN
--
Fast topology-independent and global structure alignment through an FFT-based algorithm.
Click HERE to access the web server.
 MPRDock
--
Protein-ensemble-RNA docking by efficient consideration of protein flexibility through multiple protein structures that are built by homology modeling.
MPRDock
--
Protein-ensemble-RNA docking by efficient consideration of protein flexibility through multiple protein structures that are built by homology modeling.
Click HERE to download the standalone DITScorePR scoring function.
Click HERE to download the docking decoys used in the MPRDock paper.
 PepBDB
--
a comprehensive structural database of all the biological peptide binding complexes with peptide lengths up to 50 residues in the Protein Data Bank.
PepBDB
--
a comprehensive structural database of all the biological peptide binding complexes with peptide lengths up to 50 residues in the Protein Data Bank.
Click HERE to access the database.
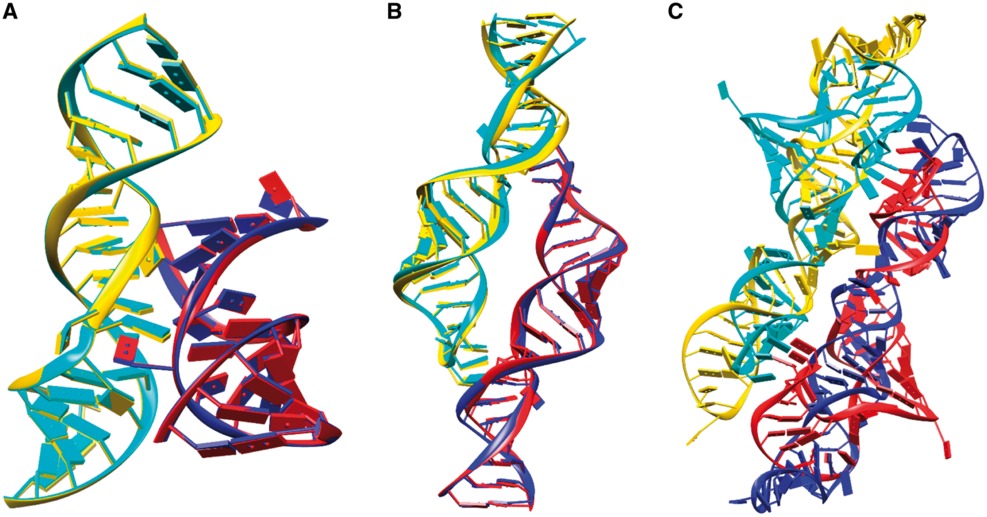 RNA Docking benchmark
-- A comprehensive and nonredundant benchmark for RNA-RNA docking and scoring.
RNA Docking benchmark
-- A comprehensive and nonredundant benchmark for RNA-RNA docking and scoring.
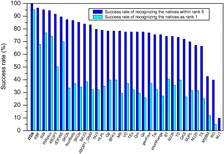 ITDA scoring function
-- A new knowledge-based scoring function with Inclusion
of Backbone Conformational Entropies from Protein Structures through backbone
dihedral angles.
ITDA scoring function
-- A new knowledge-based scoring function with Inclusion
of Backbone Conformational Entropies from Protein Structures through backbone
dihedral angles.
Click HERE to download the ITDA package.
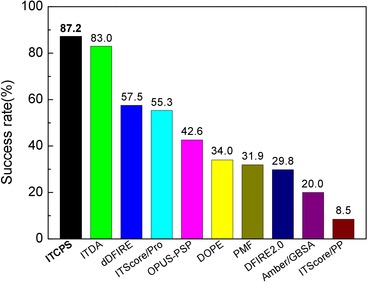 ITCPS scoring function
--
Integrating bonded and nonbonded potentials in the knowledge-based scoring function for protein structure prediction.
ITCPS scoring function
--
Integrating bonded and nonbonded potentials in the knowledge-based scoring function for protein structure prediction.
Click HERE to download the ITCPS package.