Huang Lab
About EMRNA
Deep learning based automated RNA modeling from cryo-EM maps

Copyright © 2022 Tao Li, Jiahua He, Sheng-You Huang and Huazhong University of Science and Technology Released under GNU General Public License Version 3
EMRNA is freely available for academic or commercial users. If you have any questions regarding EMRNA, please don't hesitate to contact us via huangsy@hust.edu.cn Reference:Li T, He J, Huang S-Y.* Deep learning based automated RNA modeling from cryo-EM maps. In submission, 2023 [link]
Release Notes:
2024-06-03: We have noticed EMRNA may be run failed on Ubuntu system due to the use of non-POSIX shell syntax. So we have updated EMRNA to v1.5 that should work on Ubuntu (and other Linux systems) now.2024-02-29: We have updated EMRNA to v1.4 in which gcc-6.2.0 is no longer required as we have compiled the program statically. EMRNA_v1.4 should work on CentOS 7.x or similar systems now.
In addition, per users' requests, we have also added 10 initial models named "output_*.pdb" that contains only those parts of structures with good confidences in the output directory.
2024-02-23: EMRNA v1.3 was released.
Download EMRNA
Install EMRNA
System
CentOS Linux 7.x (or other unix-based systems)External programs
EternaFold (1.3.1) (https://github.com/eternagame/EternaFold)LKH-3 (3.0.6) (http://webhotel4.ruc.dk/~keld/research/LKH-3)
These programs can be easily installed according to the documentation on the websites. We already included program "CSSR" under GNU General Public License V3.
Quick installation of python and required online packages
$ conda env create -f environment.yml$ pip install missing-packages in your EMRNA environment.
Details of required online packages
Python (3.8.8) (https://www.python.org) Python package requirements: sklearn (https://scikit-learn.org/stable/install.html) pytorch (1.9.0+cuda11.1) (https://pytorch.org) torchvision (0.10.0+cuda11.1) (https://pytorch.org) cudatoolkit (11.1) (https://developer.nvidia.com/cuda-toolkit) numpy (1.20.1) (https://www.numpy.org) einops (0.3.2) (https://einops.rocks/) mrcfile (1.3.0) (https://github.com/ccpem/mrcfile) timm(0.4.12) (https://github.com/rwightman/pytorch-image-models) tqdm (4.60.0) (https://github.com/tqdm/tqdm)NOTE: In order to run Python scripts and EMRNA properly, users should properly set the variables in EMRNA.sh :
1. Set "activate" to path of conda activator, for example
2. Set "EMRNA_env" to name of the python conda virtual environment that have all the required packages installed. An conda environment named "emrna" will be created using the quick installation command, so
3. Set "LKH_dir" to the path of LKH-3, for example
4. Set "EMRNA_home" to the path of EMRNA, for example
Please do not use path like "~/...", the Tilde "~" in the path may not be properly recongnized in EMRNA.sh
This command will generate an ELF file with name like "interp3d.cpython-*.so". Please keep "interp3d.cpython-*.so" with all python scripts "*.py" in the same directory.
It should be noted that the version of f2py should match the version of Python of the conda environment of EMRNA. Fortran compiler (e.g. gfortran, ifort, etc) is required to run f2py. For Linux systems with Debian package management (e.g. Debian, Ubuntu), gfortran can be easily installed via
Note that EMRNA is only run on linux-64 systems.
activate="/home/taoli/anaconda3/bin/activate"EMRNA_env="emrna". If the environment is built with a different name, users should modify "EMRNA_env" accordingly
3. Set "LKH_dir" to the path of LKH-3, for example
LKH_dir="/home/taoli/LKH-3.0.6"4. Set "EMRNA_home" to the path of EMRNA, for example
EMRNA_home="/home/taoli/EMRNA_v1.5/"In addition to online packages, the interpolation program "interp3d.f90" should be built as a python package 'interp3d' using f2py in the conda virtual environment of EMRNA
$ conda activate emrna
$ f2py -c ./interp3d.f90 -m interp3d$ sudo apt-get install gfortran
How to Run EMRNA
Currently, EMRNA(v1.5) is designed to build a single RNA chain for RNA-only maps.
$ /path/to/EternaFold/src/contrafold predict input_seq.fasta \
--params /path/to/EternaFold/parameters/EternaFoldParams.v1 | tail -1 > input_ss.txt ((((((..((((.........)))).(((((.......))))).....(((((.......))))))))))).....Step 2. Run EMRNA with input sequence and predicted secondary structure
$ /path/to/EMRNA_v1.5/EMRNA.sh input_map.mrc input_seq.fasta input_ss.txt out_dir [Options] Required arguments: input_map.mrc: File name of input EM density map in MRC2014 format. input_seq.fasta: File name of input sequence in fasta format. input_ss.txt: File name of input secondary structure (last line is the SS in dot-bracket representation). out_dir: Directory of the outputs (all intermediate or output files are written in this directory). Options: --contour CONTOUR: Contour level of input map, voxels below will be ignored. (default: '1e-6') -g GPU_ID: ID(s) of GPU devices to use. e.g. '0' for GPU #0, and '2,3,6' for GPUs #2, #3, and #6. (default: '0') -b BATCH_SIZE: Number of boxes input into EMRNA in one batch. (default: 40) --usecpu: Run deep learning predictin of EMRNA on CPU instead of GPU. --ncpu NCPU: Number of cpus to use to accelarate threading traces. (default: '4') Please reduce the BATCH_SIZE if CUDA runs out of memory.
Examples
$ /path/to/EternaFold/src/contrafold predict input_seq.fasta \
--params /path/to/EternaFold/parameters/EternaFoldParams.v1 | tail -1 > input_ss.txt After that, run command
$ /path/to/EMRNA_v1.5/EMRNA.sh 7p7s_D.mrc input_seq.fasta input_ss.txt output
Finally, use phenix to conduct model refinement by running the following command
$ phenix.real_space_refine 7p7s_D.mrc output/ranked_0.pdb resolution=3.0
Note that the final refinement can be done by other software and is optional for users.

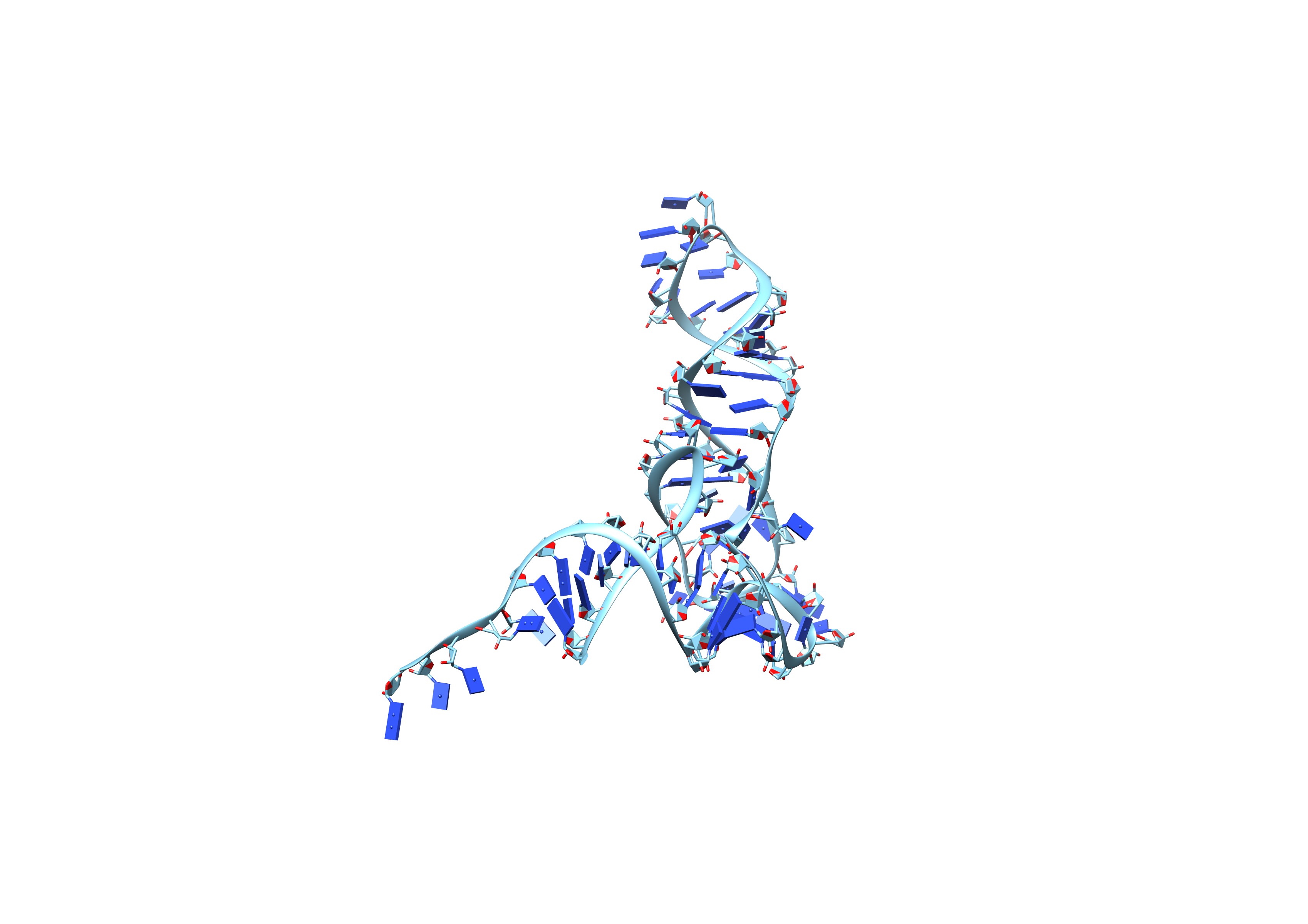
Use EMRNA to reproduce an already solved SAM-IV RNA riboswitch without any preprocessing. Try command
$ /path/to/EMRNA_v1.5/EMRNA.sh emd_20755.map input_seq.fasta input_ss.txt output -g 0 -b 40